Choose timezone
Your profile timezone:
In this example script we show how to use a Profile Cooker JSON file, the NEOTRANSP code, matlabVMEC, and the W7-X matlab tools to generate a fixed boundary VMEC input file with BEAMS3D input namelist.
Links to software
We need to define some equilirium quantities.
% Our VMEC input extension name outfile = 'W7X20180918045_cooker4700'; % % Constant value for Zeff zeff=1.271; % % NEOTRANSP needs the field on axis [T] B00axis = 2.52; % % The Profile Cooker JSON file with thomson_Te, thomson_Ti, and cxrs_Ti cooker_file='cooker_20180918045.json'; % % Now we choose which magnetic configuration we want (uncomment conf_name='EIM'; %FTM,KJM,DBM allowed
This is the main part of the script which does all the work for you.
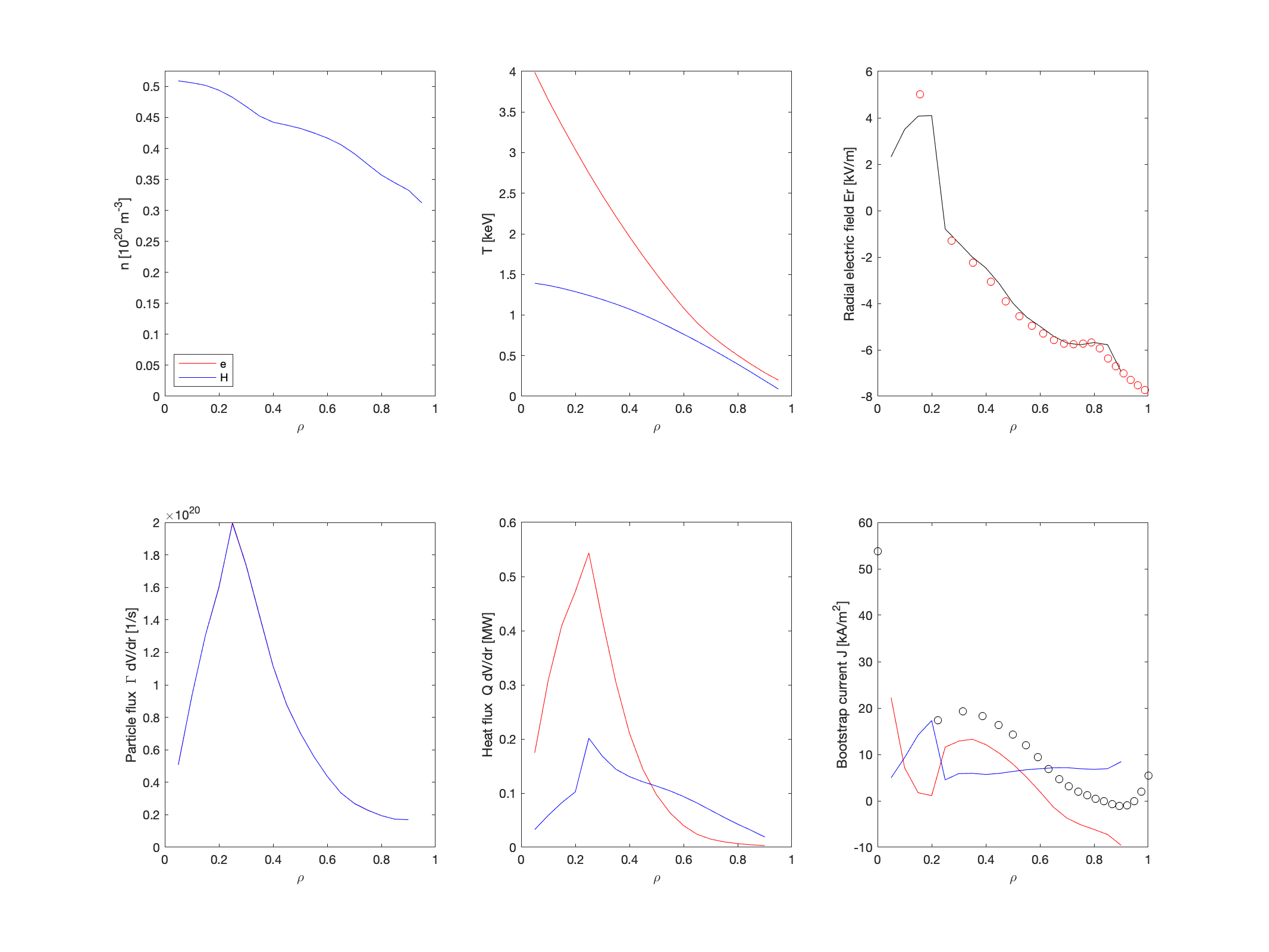
% load Json fid=fopen(cooker_file); str=fgetl(fid); fclose(fid); prof_data=jsondecode(str); % Define profile functions based on cooker data rho_vmec = 0:0.05:1; s_vmec = rho_vmec.*rho_vmec; f_te = @(s) max(pchip((prof_data.thomson_Te.reff./max(prof_data.thomson_Te.reff)).^2,... prof_data.thomson_Te.value.*1E3,s),0); f_ne = @(s) max(pchip((prof_data.thomson_ne.reff./max(prof_data.thomson_ne.reff)).^2,... prof_data.thomson_ne.value.*1E19,s),0); f_ti = @(s) max(pchip((prof_data.cxrs_Ti.reff./max(prof_data.cxrs_Ti.reff)).^2,... prof_data.cxrs_Ti.value.*1E3,s),0); dfds_te = @(s) (f_te(s+1E-6)-f_te(s-1E-6))./2E-6; dfds_ti = @(s) (f_ti(s+1E-6)-f_ti(s-1E-6))./2E-6; dfds_ne = @(s) (f_ne(s+1E-6)-f_ne(s-1E-6))./2E-6; % Setup some values based on conf_name switch conf_name case 'EIM' equilID = 'w7x-sc1'; extcur_conf=[13400*108 13400*108 13400*108 13400*108 13400*108 0*36 0*36]; case 'KJM' equilID = 'w7x-hm1'; extcur_conf=[13130*108 12760*108 12150*108 11550*108 11180*108 0*36 0*36]; case 'FTM' equilID = 'w7x-schi'; extcur_conf=[14188*108 14188*108 14188*108 14188*108 14188*108 -9790*36 -9790*36]; case 'DBM' equilID = 'w7x-scli'; extcur_conf=[11769*108 11769*108 11769*108 11769*108 11769*108 8827*36 8827*36]; otherwise disp('Please choose conf_name to be: EIM, KJM, FTM, DBM'); return; end % Define profiles for NEOTRANSP h0=0.05; rho=h0:h0:(1-h0); s=rho.*rho; Prof=[]; %Electrons Prof(1).Z=-1; Prof(1).TkeV=f_te(s).*1E-3; Prof(1).dTkeVdrho=dfds_te(s).*1E-3.*2.*rho; Prof(1).n20=f_ne(s).*1E-20; Prof(1).dn20drho=dfds_ne(s).*1E-20.*2.*rho; %Ions Prof(2).Z=1; Prof(2).m_au=1; Prof(2).TkeV=f_ti(s).*1E-3; Prof(2).dTkeVdrho=dfds_ti(s).*1E-3.*2.*rho; Prof(2).n20=f_ne(s).*1E-20; Prof(2).dn20drho=dfds_ne(s).*1E-20.*2.*rho; % Neotransp options Opt.makeErsearchplot=0; %set to 0 to turn off, or to the figure number Opt.roots='i/e'; %can be 'i', 'e', 'i&e' Opt.surfInterpVar = 'rho'; % r/a not W7AS r % Run Neotransp Transp= neotransp(equilID,rho,Prof,B00axis,Opt); % Calculate bootstrap profile and CURTOR jboot = squeeze(Transp.Bootstrapcurrdens.i)+squeeze(Transp.Bootstrapcurrdens.e)'; jboot(isnan(jboot)) = 0; jboot_spl = spline(rho,jboot); Itor = sum(jboot.*Transp.dVds'.*h0./(2.*pi.*Transp.R00')); % Calculate Phi_estatic for BEASM3D dphi=Transp.dPhidskV.*1E3; % kV to V sh = 0.5.*(s(2:end)+s(1:end-1)); %half grid f =(dphi(2:end)+dphi(1:end-1)).*0.5; %half grid phi = cumsum(f.*diff(s))+dphi(1); % Make NEOSTRANSP plot pp = pchip(sqrt(sh),phi); h3 = 0.05; s3 = 0:h3:1; rho3=sqrt(s3); plottransp(Transp); set(gcf,'Position',[1 1 1024 768],'Color','white'); drawnow; subplot(2,3,6); hold on; f=ppval(jboot_spl,rho3); plot(rho3,f./1E3,'ok'); subplot(2,3,3); hold on; f=ppval(pp,rho3); x=0.5.*(s3(2:end)+s3(1:end-1)); dphids=-diff(f)./diff(s3); Er=dphids.*sqrt(x).*2./Transp.minorradiusW7AS./1E3; plot(sqrt(x),Er,'or'); saveas(gcf,['transp_' outfile '.fig']); saveas(gcf,['transp_' outfile '.png']); close(gcf); % Write NEOTRANSP Profiles to file fid = fopen(['transp_' outfile '.txt'],'w'); fprintf(fid,'#r/a #ne [m^-3] #ni1 [m^-3] #te [eV] #ti1 [eV] #dPhi/ds [V] #jboot [A/m^2] #Gamma_e [1/s] #Gamma_i1 [1/s] #Q_e [W] #Q_i1 [W]\n'); for i=1:length(rho) fprintf(fid,'%20.10E %20.10E %20.10E %20.10E %20.10E %20.10E %20.10E %20.10E %20.10E %20.10E %20.10E\n',... rho(i),Transp.Prof(1).n20(i).*1E20,Transp.Prof(2).n20(i).*1E20,Transp.Prof(1).TkeV(i).*1E3,Transp.Prof(2).TkeV(i).*1E3,... Transp.dPhidskV(i).*1E3,Transp.Bootstrapcurrdens.i(1,1,i)+Transp.Bootstrapcurrdens.e(1,i),... Transp.Flux.e(1,i),Transp.Flux.i(1,1,i),Transp.Heatflux.e(1,i).*1E6,Transp.Heatflux.i(1,1,i).*1E6); end fclose(fid); % Setup input ec = 1.60217662E-19; p_vmec = f_ne(s_vmec).*ec.*(f_te(s_vmec)+f_ti(s_vmec)./zeff); j_vmec = ppval(jboot_spl,rho_vmec); boundary = get_w7x_fixedboundary([conf_name num2str(round(B00axis.*100),'%+3.3i')],[],[],[],[],[],[]); vmec_input = vmec_namelist_init_w7x('INDATA'); vmec_input.ns_array=[16 32 64 128]; vmec_input.ftol_array=[1E-30 1E-30 1E-30 1E-12]; vmec_input.niter_array=[1000 2000 4000 20000]; vmec_input.nstep=200; vmec_input.delt=1; vmec_input.tcon0=1; vmec_input.niter=20000; vmec_input.gamma=0; vmec_input.bloat=1; vmec_input.spres_ped=1; vmec_input.pres_scale=1; vmec_input.pmass_type='akima_spline'; vmec_input.am_aux_s=s_vmec; vmec_input.am_aux_f=p_vmec; vmec_input.ncurr=1; vmec_input.pcurr_type='akima_spline_ip'; vmec_input.curtor=Itor; vmec_input.ac_aux_s=s_vmec; vmec_input.ac_aux_f=j_vmec; vmec_input.nfp = 5; vmec_input.mpol = 13; vmec_input.ntor = 10; vmec_input.nzeta=24; vmec_input.ntheta=32; vmec_input.lfreeb=0; vmec_input.extcur=-extcur_conf; vmec_input.datatype='VMEC_input'; vmec_input.phiedge=boundary.phiedge; vmec_input.xm=boundary.xm; vmec_input.xn=boundary.xn; vmec_input.rbc=boundary.rbc; vmec_input.zbs=boundary.zbs; dexm=boundary.xm==0; vmec_input.raxis=boundary.rbc(dexm); vmec_input.zaxis=boundary.zbs(dexm); vmec_input=rmfield(vmec_input,'bcrit'); % Now write input file filename=['input.' outfile]; write_vmec_input_w7x(filename,vmec_input); w7x_beam_params(1:8,'H2','write_beams3d','TE',[s_vmec;f_te(s_vmec)],... 'NE',[s_vmec;f_ne(s_vmec)],'TI',[s_vmec;f_ti(s_vmec)],... 'ZEFF',[s_vmec; 0.*s_vmec+zeff],... 'POT',[s3;ppval(pp,rho3)],'file',filename,'t_end',0.100,... 'NPOINC',3,'nphi',72);
The equilibrium w7x-sc1 is in the database. Taking the DKES data from there.
Performing Er search
rho=0.050, residule: e0.0047, : iInf, e0.0047
rho=0.100, residule: e0.0142, e0.0002, : iInf, e0.0002
rho=0.150, residule: i0.1411, e0.0150, i0.1240, e0.0016, i0.1538, e0.0000, i0.0801, e0.0000, i0.0513, e0.0000, i0.0351, e0.0000, i0.0249, e0.0000, i0.0181, e0.0000, i0.0134, e0.0000, i0.0100, e0.0000, i0.1585, e0.0008, i0.0882, e0.0004, i0.0399, e0.0002, i0.0101, e0.0001, i0.0000, e0.0000, : i0.0000, e0.0000
rho=0.200, residule: i0.0150, e0.0302, i0.0022, e0.0027, : i0.0022, e0.0027
rho=0.250, residule: i0.0808, i0.0024, : i0.0024, eInf
rho=0.300, residule: i0.0823, i0.0119, i0.0018, : i0.0018, eInf
rho=0.350, residule: i0.0559, i0.0105, i0.0025, : i0.0025, eInf
rho=0.400, residule: i0.0352, i0.0074, i0.0014, : i0.0014, eInf
rho=0.450, residule: i0.0202, i0.0015, : i0.0015, eInf
rho=0.500, residule: i0.0135, i0.0002, : i0.0002, eInf
rho=0.550, residule: i0.0057, i0.0000, : i0.0000, eInf
rho=0.600, residule: i0.0108, i0.0001, : i0.0001, eInf
rho=0.650, residule: i0.0104, i0.0001, : i0.0001, eInf
rho=0.700, residule: i0.0016, : i0.0016, eInf
rho=0.750, residule: i0.0035, : i0.0035, eInf
rho=0.800, residule: i0.0222, i0.0003, : i0.0003, eInf
rho=0.850, residule: i0.0440, i0.0005, : i0.0005, eInf
rho=0.900, residule: i0.0390, i0.0005, : i0.0005, eInf
rho=0.950, residule:
Warning: All NaN entries in the first and second input were ignored.
Configuration: EIM+252